
University of Washington and Microsoft scientists have introduced a new class of reporter proteins that can be immediately read by a commercially available nanopore sensing system. Here, UW doctoral pupil Nicolas Cardozo pipettes a remedy made up of NanoporeTERs onto a portable MinION machine as UW research assistant professor Jeff Nivala appears to be like on. Credit rating: Dennis Wise/College of Washington
Genetically encoded reporter proteins have been a mainstay of biotechnology research, allowing scientists to monitor gene expression, have an understanding of intracellular procedures, and debug engineered genetic circuits.
But traditional reporting schemes that count on fluorescence and other optical approaches arrive with simple limitations that could cast a shadow around the field’s future progress. Now, scientists at the College of Washington and Microsoft have designed a “nanopore-tal” into what is going on within these sophisticated biological devices, making it possible for researchers to see reporter proteins in a whole new light.
The team introduced a new course of reporter proteins that can be right study by a commercially accessible nanopore sensing machine. The new program — dubbed “Nanopore-addressable protein Tags Engineered as Reporters” or “NanoporeTERs” — can detect multiple protein expression stages from bacterial and human cell cultures considerably further than the ability of current methods.
The examine was posted on August 12, 2021, in Mother nature Biotechnology.

College of Washington and Microsoft scientists have launched a new course of reporter proteins that can be right go through by a commercially obtainable nanopore sensing unit. Raw nanopore alerts stream from the MinION product, which includes an array of hundreds of nanopore sensors. Each and every color signifies knowledge from an individual nanopore. The crew employs device understanding to interpret these alerts as NanoporeTERs barcodes. Credit score: Dennis Smart/College of Washington
“NanoporeTERs offer you a new and richer lexicon for engineered cells to categorical them selves and shed new light-weight on the things they are developed to monitor. They can tell us a good deal a lot more about what is going on in their atmosphere all at when,” said co-guide writer Nicolas Cardozo, a doctoral scholar with the UW Molecular Engineering and Sciences Institute. “We’re primarily making it doable for these cells to ‘talk’ to desktops about what’s happening in their surroundings at a new stage of depth, scale and effectiveness that will allow deeper assessment than what we could do in advance of.”
For common labeling approaches, researchers can keep track of only a couple optical reporter proteins, these kinds of as green fluorescent protein, concurrently due to the fact of their overlapping spectral attributes. For instance, it’s tricky to distinguish amongst a lot more than a few different colors of fluorescent proteins at as soon as. In distinction, NanoporeTERs have been designed to have unique protein “barcodes” composed of strings of amino acids that, when used in mixture, let at minimum 10 periods more multiplexing possibilities.
These artificial proteins are secreted outside the house of a cell into the encompassing atmosphere, the place researchers can obtain and evaluate them utilizing a commercially accessible nanopore array. In this article, the crew applied the Oxford Nanopore Systems MinION product.
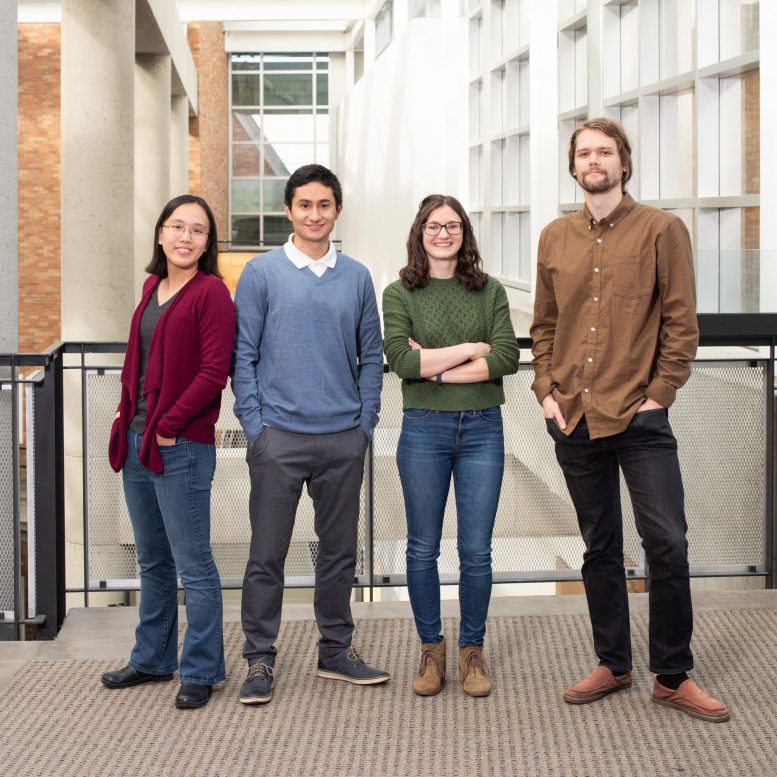
University of Washington and Microsoft researchers have introduced a new course of reporter proteins that can be specifically read through by a commercially obtainable nanopore sensing system. Demonstrated here (from remaining to appropriate): Karen Zhang, Nicolas Cardozo, Kathryn Doroschak and Jeff Nivala. Not pictured: Aerilynn Nguyen, Zoheb Siddiqui, Nicholas Bogard, Karin Strauss and Luis Ceze. Observe: this picture was taken right before the COVID-19 pandemic. Credit history: Tara Brown Images
The researchers engineered the NanoporeTER proteins with charged “tails” so that they can be pulled into the nanopore sensors by an electric powered area. Then the crew utilizes machine understanding to classify the electrical signals for just about every NanoporeTER barcode in purchase to ascertain each individual protein’s output degrees.
“This is a essentially new interface in between cells and personal computers,” explained senior writer Jeff Nivala, a UW study assistant professor in the Paul G. Allen University of Laptop Science & Engineering. “One analogy I like to make is that fluorescent protein reporters are like lighthouses, and NanoporeTERs are like messages in a bottle.
“Lighthouses are genuinely beneficial for speaking a bodily place, as you can pretty much see wherever the sign is coming from, but it is tricky to pack more information into that type of sign. A concept in a bottle, on the other hand, can pack a ton of information into a extremely smaller vessel, and you can ship lots of of them off to an additional site to be browse. You may possibly drop sight of the specific actual physical locale where by the messages were despatched, but for quite a few applications that is not going to be an issue.”
As a proof of thought, the crew formulated a library of far more than 20 distinct NanoporeTERs tags. But the opportunity is appreciably bigger, according to co-guide writer Karen Zhang, now a doctoral student in the UC Berkeley-UCSF bioengineering graduate method.
“We are currently working to scale up the number of NanoporeTERs to hundreds, 1000’s, maybe even millions extra,” reported Zhang, who graduated this calendar year from the UW with bachelor’s degrees in each biochemistry and microbiology. “The a lot more we have, the far more issues we can track.
“We’re specifically energized about the probable in one-cell proteomics, but this could also be a activity-changer in conditions of our capacity to do multiplexed biosensing to diagnose illness and even target therapeutics to specific locations within the human body. And debugging difficult genetic circuit layouts would become a entire great deal simpler and significantly significantly less time-consuming if we could evaluate the performance of all the elements in parallel instead of by trial and mistake.”
These scientists have built novel use of the MinION device just before, when they formulated a molecular tagging process to exchange typical inventory handle techniques. That system relied on barcodes comprising synthetic strands of DNA that could be decoded on demand from customers using the moveable reader.
This time, the crew went a action even more.
“This is the 1st paper to exhibit how a business nanopore sensor product can be repurposed for programs other than the DNA and RNA sequencing for which they have been originally developed,” said co-writer Kathryn Doroschak, a computational biologist at Adaptive Biotechnologies who finished this get the job done as a doctoral scholar at the Allen Faculty. “This is fascinating as a precursor for nanopore technologies becoming a lot more obtainable and ubiquitous in the potential. You can previously plug a nanopore device into your mobile cellular phone. I could envision someday owning a option of ‘molecular apps’ that will be comparatively low-cost and widely obtainable outside the house of common genomics.”
Reference: “Multiplexed direct detection of barcoded protein reporters on a nanopore array” by Nicolas Cardozo, Karen Zhang, Kathryn Doroschak, Aerilynn Nguyen, Zoheb Siddiqui, Nicholas Bogard, Karin Strauss, Luis Ceze and Jeff Nivala, 12 August 2021, Nature Biotechnology.
DOI: 10.1038/s41587-021-01002-6
Additional co-authors of the paper are Aerilynn Nguyen at Northeastern University and Zoheb Siddiqui at Amazon, both of those former UW undergraduate students Nicholas Bogard at Patch Biosciences, a previous UW postdoctoral exploration associate Luis Ceze, an Allen College professor and Karin Strauss, an Allen School affiliate professor and a senior principal exploration manager at Microsoft. This investigation was funded by the Countrywide Science Basis, the Nationwide Institutes of Health and fitness and a sponsored research agreement from Oxford Nanopore Technologies.




More Stories
Mutual Fund Selection for Advanced Traders: Factor Tilts, Style Drift Detection, and Performance Persistence Analytics
How a SaaS Marketing Agency Can Accelerate Your Software Growth
Why AI Is the Key to Running an Organized Data Ecosystem